Phylogenomics
"Nothing in biology makes sense except in the light of evolution" - Theodosius Dobzhansky.
As an evolutionary and comparative biologist, all my work begins with a phylogeny (a tree of life-style diagram that represents the relationships between lineages). The 21st century has seen an explosion of methods and technologies for collecting genomic data that allow us to ask and address increasingly precise and complex questions but also require equally rigorous and complicated analyses. My work frequently involves developing or applying new phylogenomic methods, from cleaning and assembling data to novel data integration strategies and comparative analyses.
Merging phylogenomic data and the evolution of photosynthetic anatomy
Many types of phylogenomic data are collected but rarely combined. We developed a framework to do this to increase sampling breadth and density in the Portullugo (Caryophyllales) in order to analyze the co-evolution of photosynthetic anatomy and physiology.
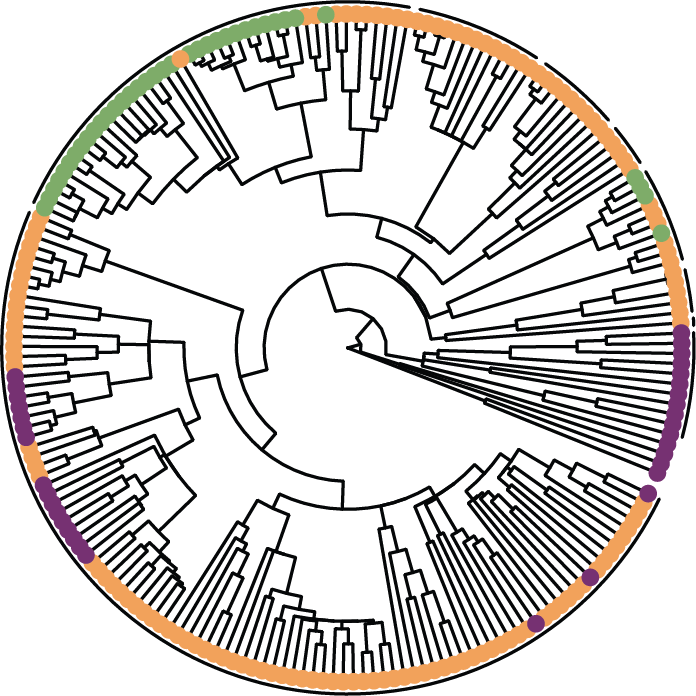
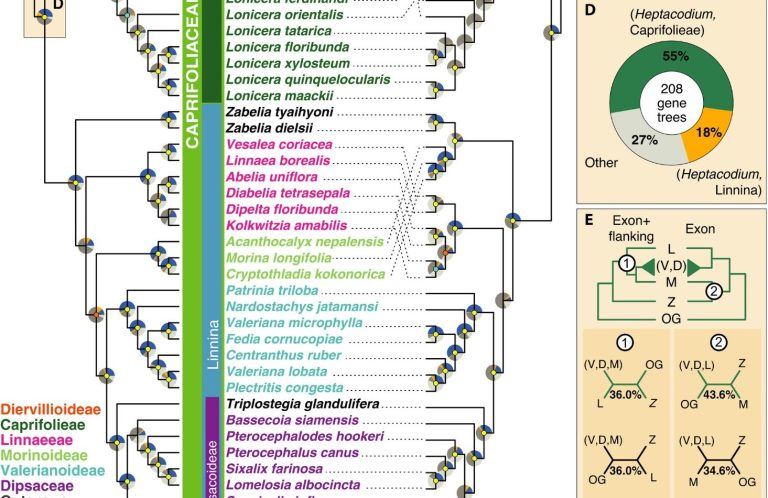
Target enrichment and phylogenetic hypothesis testing
In one of the first applications of the Angiosperm353 Probe Set, we reconstructed an updated phylogeny for the Dipsacales and tested decades-old hypotheses about ancient hybridization.
Genomics from museum specimens
Museums and herbaria hold hundreds of years of biological records that provide rich information about species' historical ranges and even extinct lineages. Using a reduced representation technique, we were able to reconstruct the evolutionary history of a rare holoparasitic lineage from herbarium specimens.
©Copyright. All rights reserved.
We need your consent to load the translations
We use a third-party service to translate the website content that may collect data about your activity. Please review the details in the privacy policy and accept the service to view the translations.